featsel_ex1
Examples of various feature selection procedures, organised per procedure
PRTools and PRDataSets should be in the path
Download the m-file from here. See http://37steps.com/prtools for more.
Feature curves are shown for 6 feature rankings computed by 3 procedures:
- Individual selection
- Forward selection
- Backward selecton
and 2 criteria:
- The Mahalanobis distance
- The leave-one-out nearest neighbor performance
These criteria are computed for the entire training set. Each of the 6 plots shows the performance of 3 classifiers based on a 50-50 random split of the dataset for training and testing.
- 1-NN rule
- The Fisher classifier
- The linear support vector machine.
In another, very similar example, featsel_ex2 the same curves are organised per classifier instead of per ranking procedure.
Contents
- Show dataset
- Define classifiers
- Original feature curves
- Individual selection by the Mahalanobis criterion
- Forward selection by the Mahalanobis criterion
- Backward selection by the Mahalanobis criterion
- Individual selection by the NN criterion
- Forward selection by the NN criterion
- Backward selection by the NN criterion
Show dataset
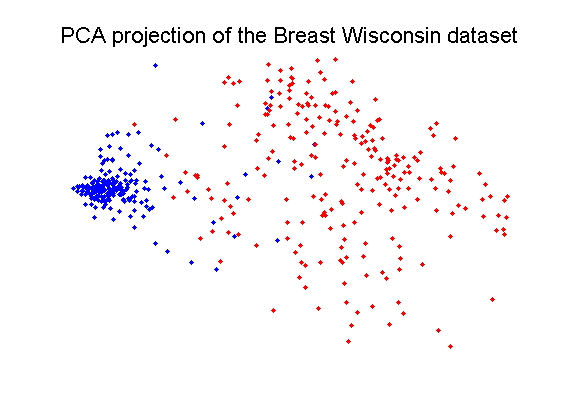
The Breast Wisconsin dataset is based on 9 features and 683 objects in two classes of 444 and 239 objects.
delfigs a = breast; a = setprior(a,0); scattern(a*pcam(a,2)); title(['PCA projection of the ' getname(a) ' dataset'])
Define classifiers
w1 = setname(knnc([],1),'1-NN'); w2 = setname(fisherc,'Fisher'); w3 = setname(libsvc,'LibSVC-1'); % number of random train-test splits used in the feature curve routine. nreps = 25;
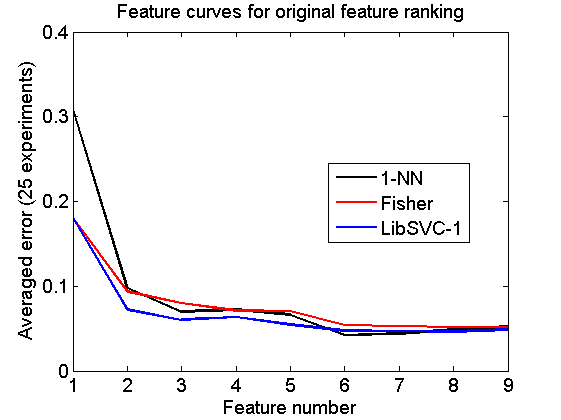
Original feature curves
These curves are based on the feature ranking of the original dataset
randreset; e = clevalf(a,{w1,w2,w3},[],0.5,nreps);
figure; plote(e);
title('Feature curves for original feature ranking')
xlabel('Feature number'); set(gca,'xscale','linear');
set(gca,'xticklabel',1:size(a,2)); set(gca,'xtick',1:size(a,2));
fontsize(14);
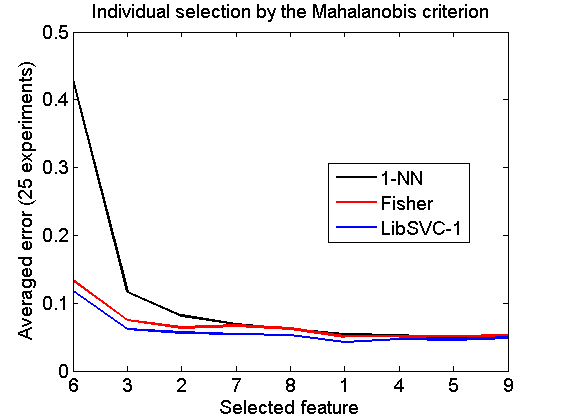
Individual selection by the Mahalanobis criterion
v = featseli(a,'maha-s',size(a,2)); randreset; e = clevalf(a*v,{w1,w2,w3},[],0.5,nreps); figure; plote(e); title('Individual selection by the Mahalanobis criterion') xlabel('Selected feature'); set(gca,'xscale','linear'); set(gca,'xticklabel',+v); set(gca,'xtick',1:size(a,2)); fontsize(14);
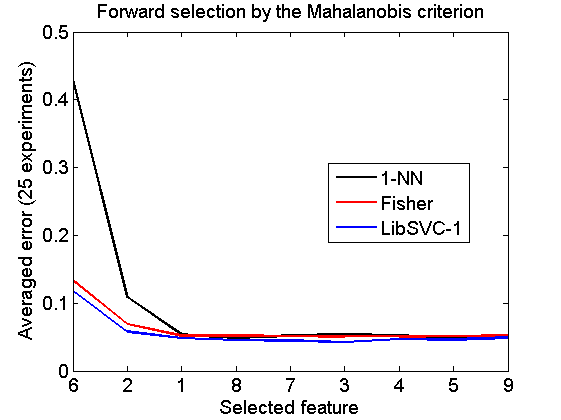
Forward selection by the Mahalanobis criterion
v = featself(a,'maha-s',size(a,2)); randreset; e = clevalf(a*v,{w1,w2,w3},[],0.5,nreps); figure; plote(e); title('Forward selection by the Mahalanobis criterion') xlabel('Selected feature'); set(gca,'xscale','linear'); set(gca,'xticklabel',+v); set(gca,'xtick',1:size(a,2)); fontsize(14);
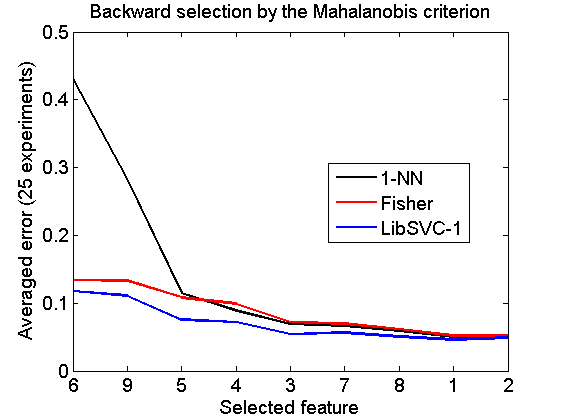
Backward selection by the Mahalanobis criterion
[v,r] = featselb(a,'maha-s',1); % the next statment is needed to retrieve the feature ranking v = featsel(size(a,2),[+v abs(r(2:end,3))']); randreset; e = clevalf(a*v,{w1,w2,w3},[],0.5,nreps); figure; plote(e); title('Backward selection by the Mahalanobis criterion') xlabel('Selected feature'); set(gca,'xscale','linear'); set(gca,'xticklabel',+v); set(gca,'xtick',1:size(a,2)); fontsize(14);
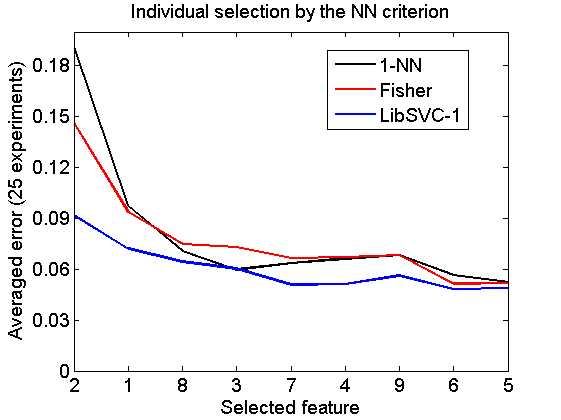
Individual selection by the NN criterion
v = featseli(a,'NN',size(a,2)); randreset; e = clevalf(a*v,{w1,w2,w3},[],0.5,nreps); figure; plote(e); title('Individual selection by the NN criterion') xlabel('Selected feature'); set(gca,'xscale','linear'); set(gca,'xticklabel',+v); set(gca,'xtick',1:size(a,2)); fontsize(14);
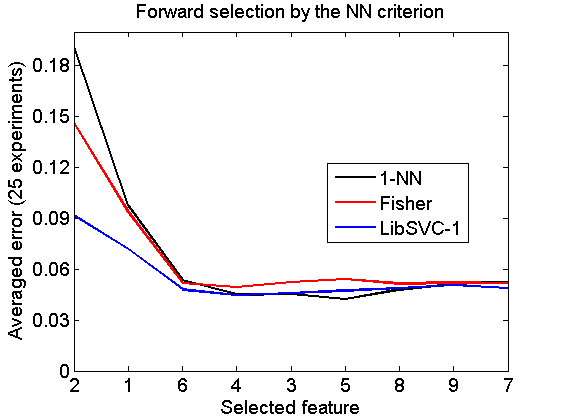
Forward selection by the NN criterion
v = featself(a,'NN',size(a,2)); randreset; e = clevalf(a*v,{w1,w2,w3},[],0.5,nreps); figure; plote(e); title('Forward selection by the NN criterion') xlabel('Selected feature'); set(gca,'xscale','linear'); set(gca,'xticklabel',+v); set(gca,'xtick',1:size(a,2)); fontsize(14);
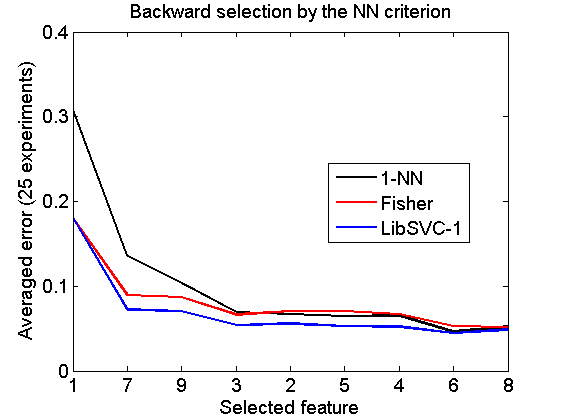
Backward selection by the NN criterion
[v,r] = featselb(a,'NN',1); % the next statment is needed to retrieve the feature ranking v = featsel(size(a,2),[+v abs(r(2:end,3))']); randreset; e = clevalf(a*v,{w1,w2,w3},[],0.5,nreps); figure; plote(e); title('Backward selection by the NN criterion') xlabel('Selected feature'); set(gca,'xscale','linear'); set(gca,'xticklabel',+v); set(gca,'xtick',1:size(a,2)); fontsize(14);